import networkx as nx
import itertools
import matplotlib.pyplot as plt
import scipy
# Define the possible nucleotides
nucleotides = ['A', 'C', 'G', 'T']
# Generate all possible DNA sequences of length 3
sequences = [''.join(seq) for seq in itertools.product(nucleotides, repeat=3)]
# Initialize an empty graph
G = nx.Graph()
# Add nodes for each possible sequence
G.add_nodes_from(sequences)
# Function to compute Hamming distance
def hamming_distance(seq1, seq2):
return sum(c1 != c2 for c1, c2 in zip(seq1, seq2))
# Add edges between nodes with Hamming distance of 1
for i in range(len(sequences)):
for j in range(i + 1, len(sequences)):
if hamming_distance(sequences[i], sequences[j]) == 1:
G.add_edge(sequences[i], sequences[j])
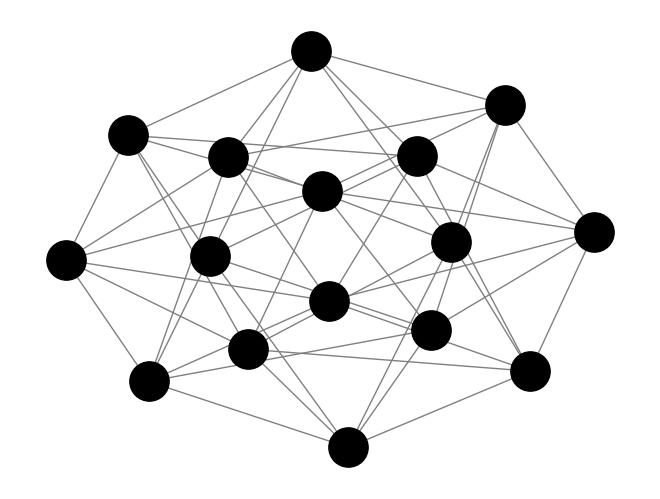
# Draw the graph
pos = nx.kamada_kawai_layout(G) # positions for all nodes
nx.draw(G, pos, with_labels=True, node_color='black', edge_color='gray', node_size=400, font_size=10)
# Display the plot
plt.savefig("graph.svg")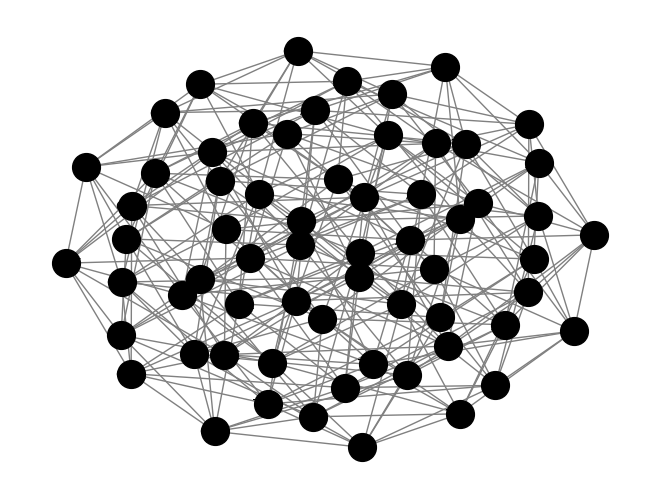
# Define the possible nucleotides
nucleotides = ['A', 'C', 'G', 'T']
# Generate all possible DNA sequences of length 3
sequences = [''.join(seq) for seq in itertools.product(nucleotides, repeat=2)]
# Initialize an empty graph
G = nx.Graph()
# Add nodes for each possible sequence
G.add_nodes_from(sequences)
# Function to compute Hamming distance
def hamming_distance(seq1, seq2):
return sum(c1 != c2 for c1, c2 in zip(seq1, seq2))
# Add edges between nodes with Hamming distance of 1
for i in range(len(sequences)):
for j in range(i + 1, len(sequences)):
if hamming_distance(sequences[i], sequences[j]) == 1:
G.add_edge(sequences[i], sequences[j])# Draw the graph
pos = nx.spring_layout(G) # positions for all nodes
nx.draw(G, pos, with_labels=True, node_color='grey', edge_color='gray', node_size=400, font_size=10)
# Display the plot
plt.savefig("graph2.svg")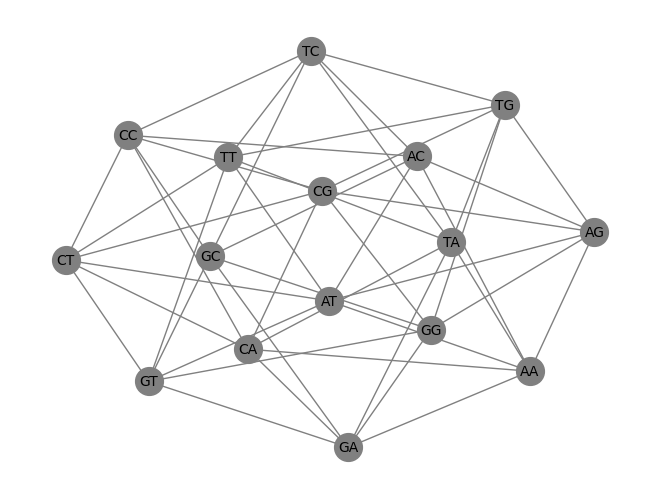
nx.draw(G, pos, with_labels=True, node_color='black', edge_color='gray', node_size=800, font_size=11)
plt.savefig("graph2.svg")